ML4360 - Computer vision
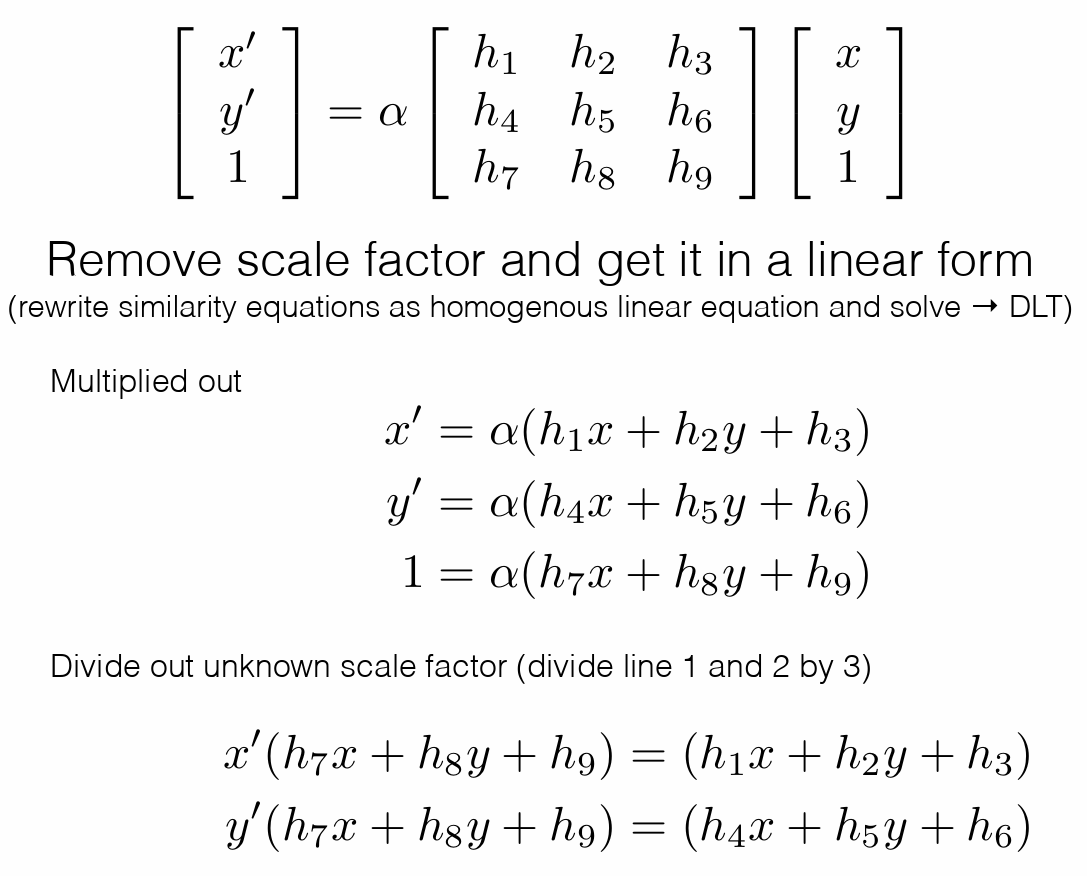
Direct Linear Transform
Concatenate \(A_{i}\) into single \(2n\times 9\) matrix \(A\) leads to the following constrained least squares problem
where we have fixed \(||\tilde{h}||_{2}^{2}=1\) as \(\tilde{H}\) is is homogeneous (i.e., defined only up to scale).
The solution to the above optimization problem is the singular vector corresponding to the smallest singular value of A. The resulting algorithm is called Direct Linear Transformation.
Singular Value Decomposition
The SVD of \(m \times n\) matrix \(A\) is given by \(A=U\Sigma V^{T}=\sum_{i=1}^{r}u_{i}\sigma _{i}v_{i}^{T}\)
where:
-
\(U\): \(m\times m\) orthogonal matrix of the orthonormal eigenvectors of \(AA^{T}\)
-
\(V^{T}\): transpose of a \(n\times n\) orthogonal matrix containing the orthonormal eigenvectors of \(A^{T}A\)
-
\(\Sigma\): a \(m\times n\) matrix, actually a diagonal matrix with \(r\) elements equal to the root of the positive eigenvalues of \(AA^{T}\) or \(A^{T}A\) (both matrics have the same positive eigenvalues anyway) and \(m-r\) extra zero rows and \(n-r\) new zero columns.
Pseudoinverse Matrix
Definition
For \(A \in \mathbb{K}^{m\times n}\), a pseudoinverse of \(A\) is defined as a matrix \({\displaystyle A^{+}\in \mathbb {K} ^{n\times m}}\) satisfying all of the following four criteria, known as the Moore–Penrose conditions:
- \({\displaystyle AA^{+}}\) need not be the general identity matrix, but it maps all column vectors of A to themselves:
- \({\displaystyle A^{+}}\) acts like a weak inverse:
- \({\displaystyle AA^{+}}\) is Hermitian. Hermitian matrix is a complex square matrix that is equal to its own conjugate transpose.
- \({\displaystyle A^{+}A}\) is also Hermitian.
Linear least-squares
The pseudoinverse provides a least squares solution to a system of linear equations.For \({\displaystyle A\in \mathbb {K} ^{m\times n}}\), given a system of linear equations \(Ax=b\), in general, a vector \({\displaystyle x}\) that solves the system may not exist, or if one does exist, it may not be unique. More specifically, a solution exists if and only if \({\displaystyle b}\) is in the image of \({\displaystyle A}\), and is unique if and only if \({\displaystyle A}\) is injective. The pseudoinverse solves the "least-squares" problem as follows:
-
\(\forall x \in \mathbb{K}^{n}\), we want to find a \(z\) satisfying \(\Vert{Az-b}\Vert_{2}\leq \Vert{Ax-b}\Vert_{2}\), where \(z=A^{+}b\) and \(\Vert \cdot \Vert_{2}\) denotes the Euclidean norm.
-
This weak inequality holds with equality if and only if \({\displaystyle x=A^{+}b+\left(I-A^{+}A\right)w}\) for any vector \({\displaystyle w}\); this provides an infinitude of minimizing solutions unless \({\displaystyle A}\) has full column rank, in which case \({\displaystyle \left(I-A^{+}A\right)}\) is a zero matrix.
Assume we want to solve \(n\) in \(Sn=I\) where S is a real matrix. A solution may not unique. We can use pseudoinverse to solve it.
\((S^{T}S)^{-1}S^{T}\) here is pesudoinverse. * Reference: wikipedia
Chamfer Distance
Chamfer Distance is widely used in 3D reconstruction to judge how close one point cloud is on average to the other.
Given two point clouds \(X\) and \(Y\), Chamfer Distance is defined as follows:
In the following code, we use the KDTree to get the nearest neighbors of one point cloud to the other.
def chamfer_distance(pcl_0, pcl_1): # pcl_0 and pcl_1 are two point clouds in 3D space
assert pcl_1.shape[-1] == 3
assert pcl_0.shape[-1] == 3
kd_pcl_0 = KDTree(pcl_0)
kd_pcl_1 = KDTree(pcl_1)
mindis_x2y, nearest_x2y= kd_pcl_1.query(pcl_0)
mindis_y2x, nearest_y2x= kd_pcl_0.query(pcl_1)
chamfer_dist = 0.5 * float(mindis_x2y.mean() + mindis_y2x.mean())
assert type(chamfer_dist) == float
return chamfer_dist